Protenix & PXDesign:
Predict, Generate, Discover.
The Protenix Suite is a unified, open-source ecosystem that
transforms high-accuracy structural knowledge into functional design.
Protenix provides the foundation for SOTA molecular
structure prediction. This foundation powers PXDesign,
a specialized generative suite that utilizes Protenix for rigorous candidate filtering,
achieving experimental success with 17-82% nanomolar hits and
2-6× gains over strong baselines in de novo protein binder design.
Protenix: High-Accuracy Structure Prediction
Protenix is a foundation model for molecules, providing SOTA accuracy in predicting diverse molecular structures (protein, DNA, RNA, ligand, etc). As an open-source model, it is fully trainable and capable of predicting a wide range of biomolecular complexes with benchmark-level accuracy.

PXDesign: High Hit-Rate Binder Generation
PXDesign generates novel protein binders with high experimental success rates: 17-82% hit rates (KD < 1000 nM) on diverse targets including IL-7RA, PD-L1, VEGF-A, SC2RBD, TrkA, EGFR, etc.
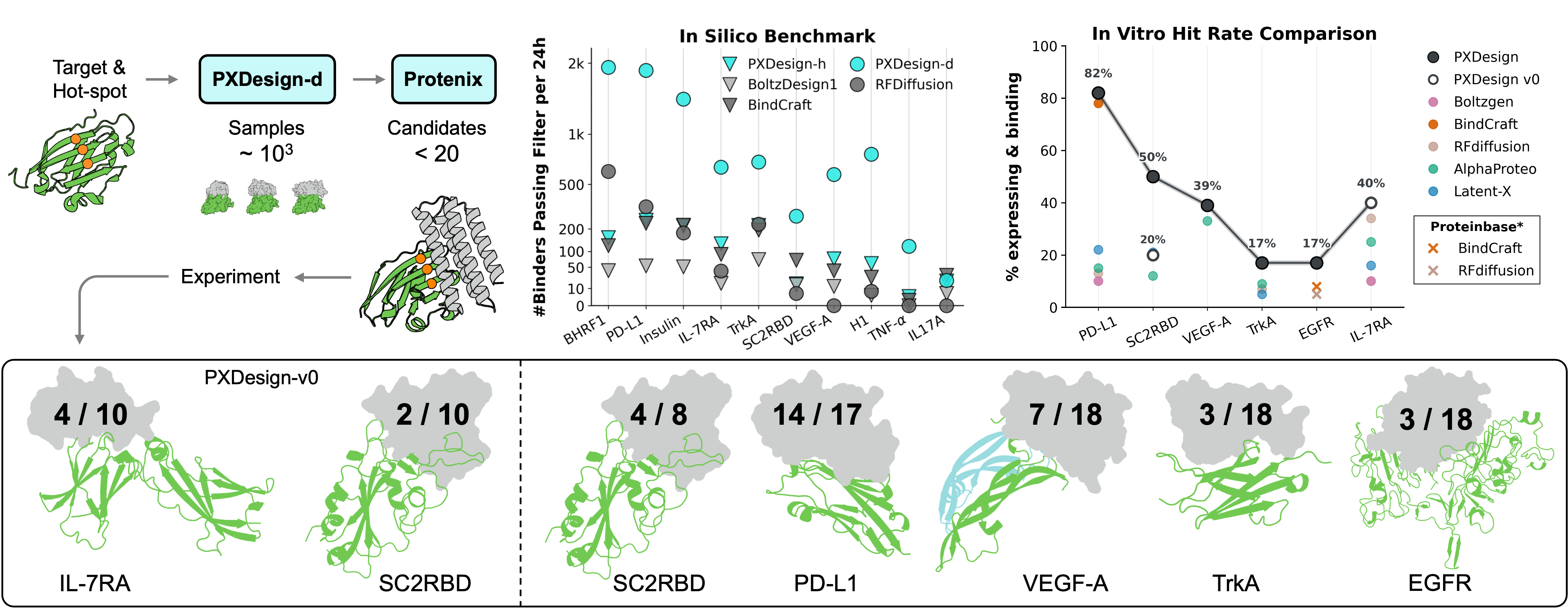
1) Generation
- PXDesign-d (Diffusion) is a DiT-style model for backbone generation — proposes many target-conditioned binder backbones; sequences are then assigned with ProteinMPNN.
2) Prediction & filtering
- Complex structures are predicted with Protenix. Candidates are filtered by confidence scores (e.g., ipTM) and structure consistency using Protenix and AF2-IG.
3) Selection for wet-lab
- Passing designs are clustered by structural similarity (Foldseek) to preserve diversity; within each cluster, higher-confidence designs (Protenix ipTM) are prioritized for expression and BLI affinity assays.
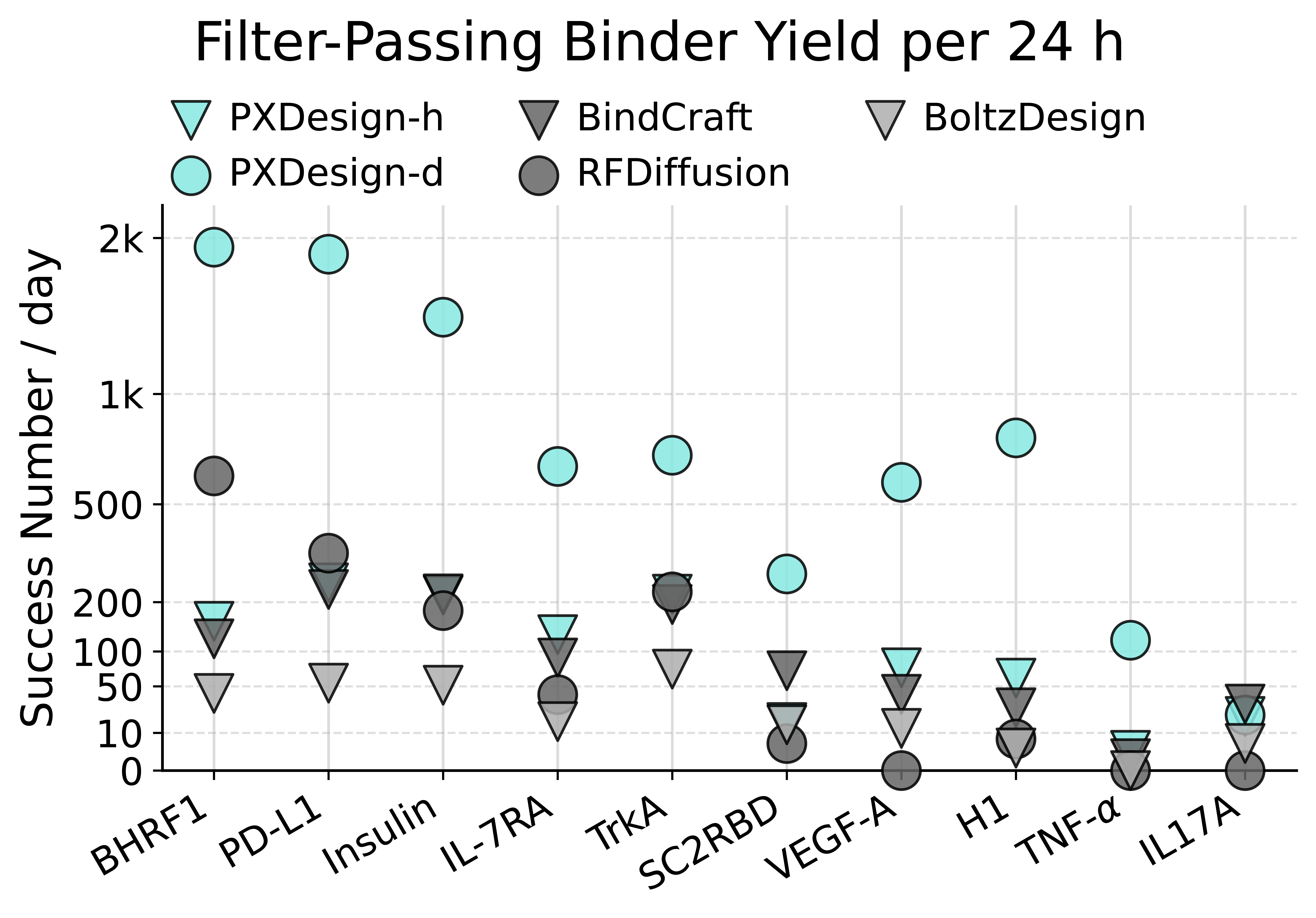
Performance Benchmarks
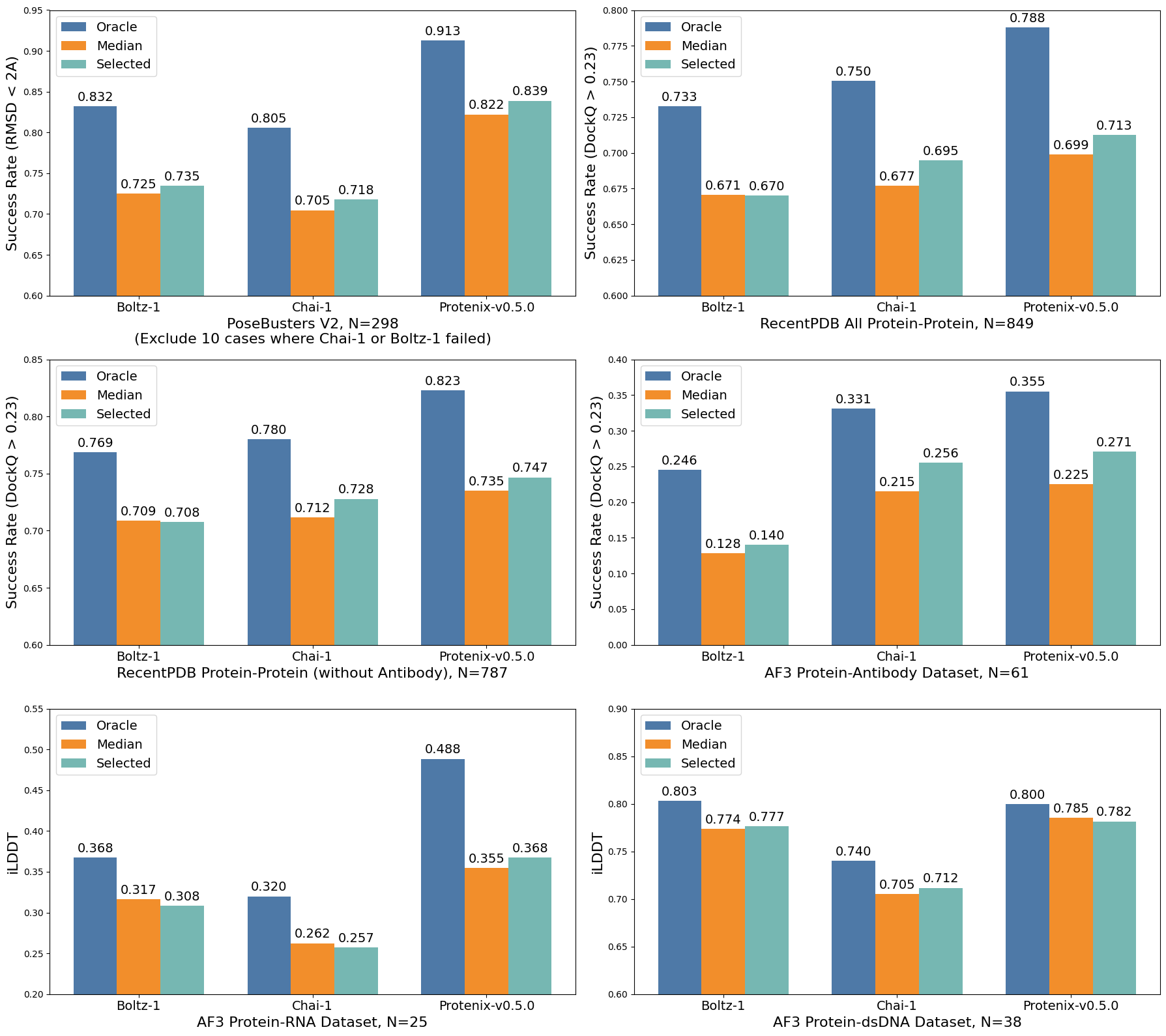
1) Structure Prediction Accuracy
Protenix achieves industry-leading accuracy on modern benchmarks, often surpassing models like Boltz-1 and Chai-1.
2) Wet-lab Validation of De Novo Binders
PXDesign achieves high nanomolar hit rates, leading or matching the best on multiple targets.
Download our designs (.zip)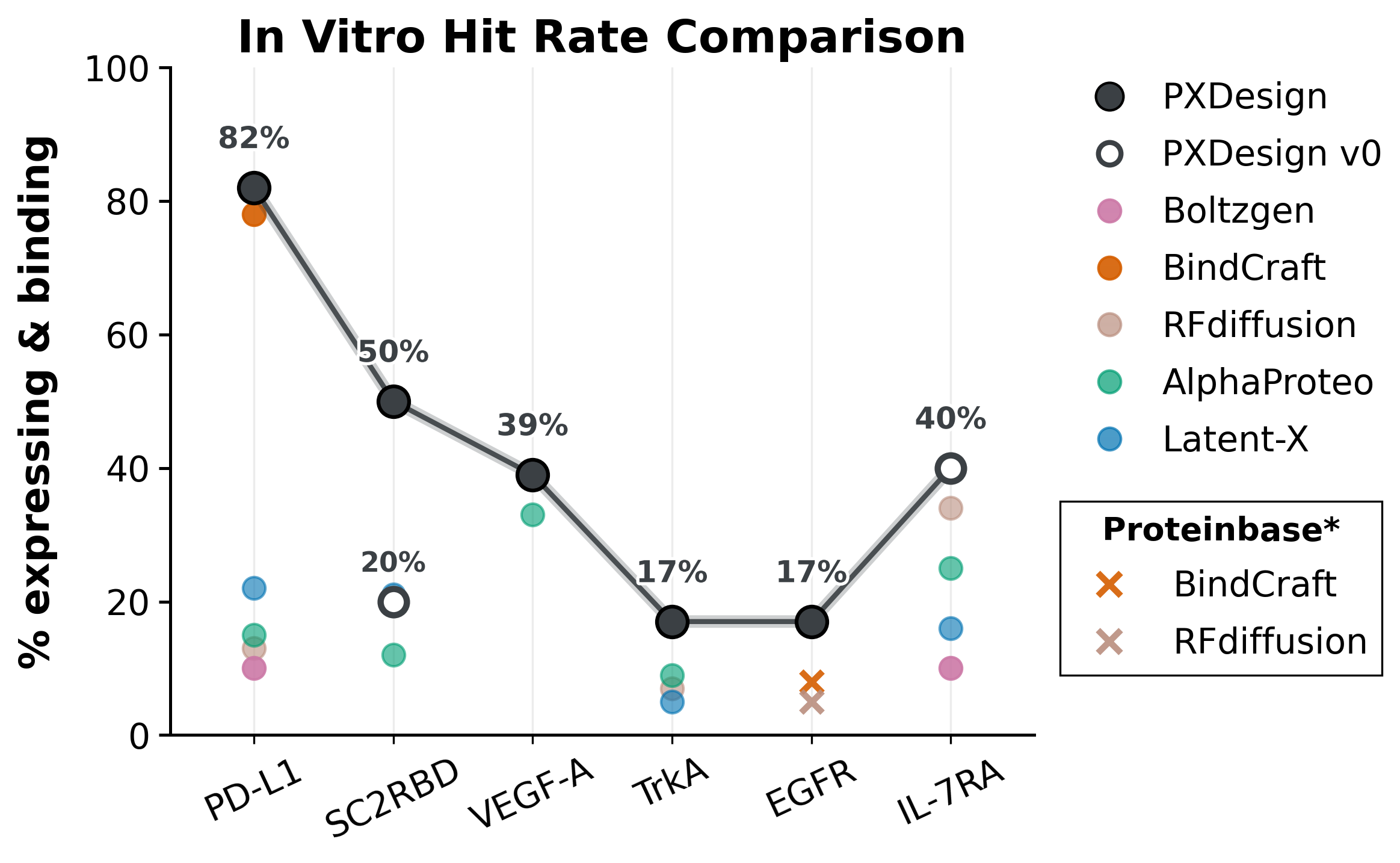
Per-target experimental success rates across methods.
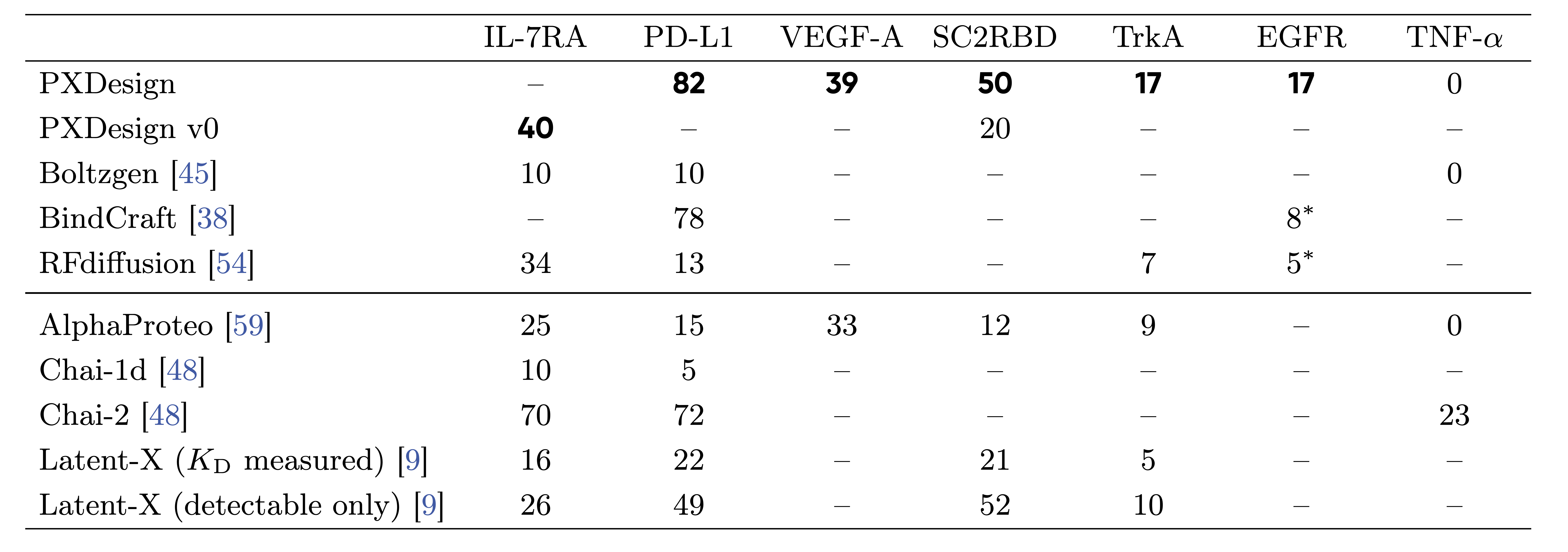
Experimental hit rates (% expressing & binding) for designed binders. “–” = not tested.
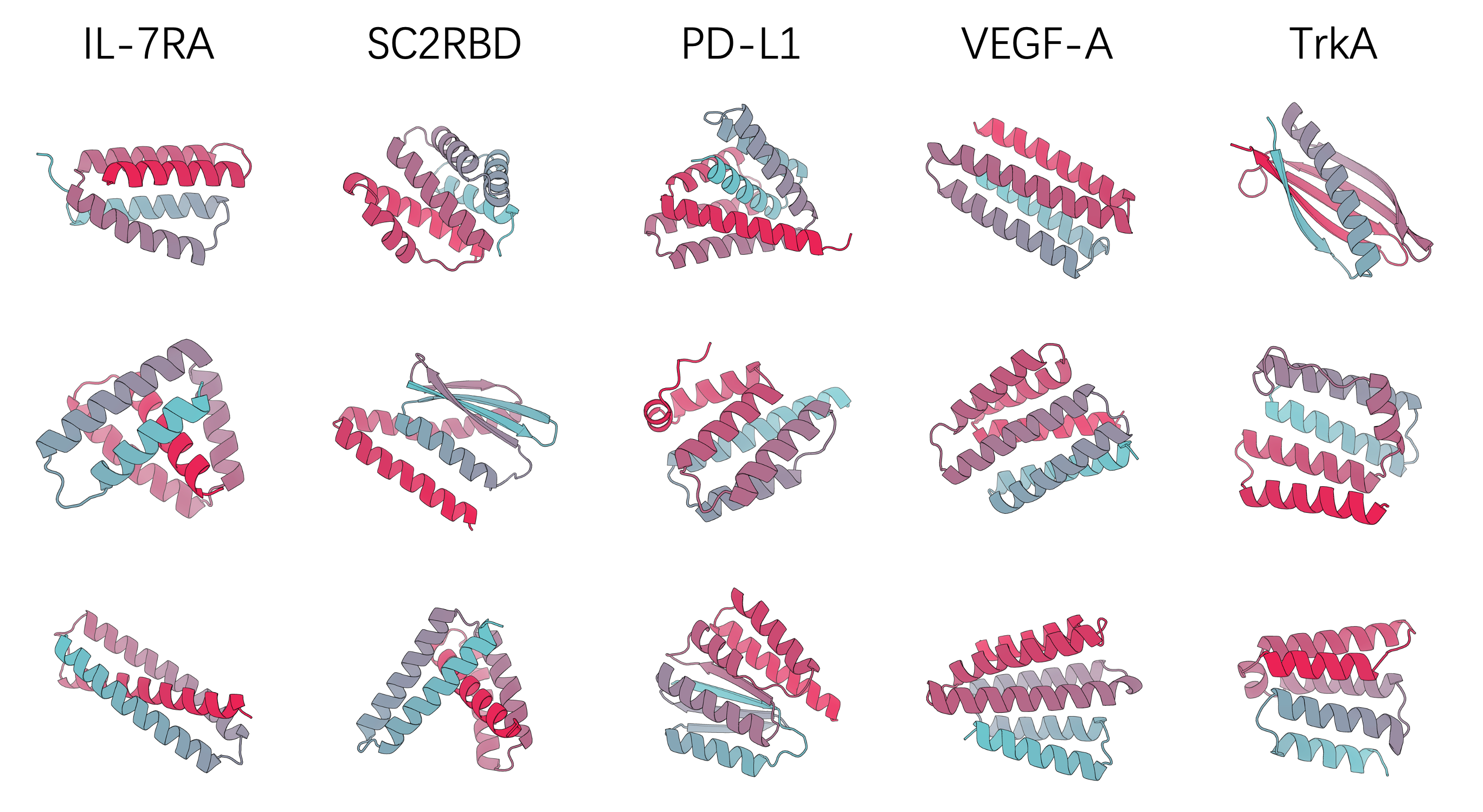
Representative PXDesign-designed nanomolar binders.
3) Filtering & Ranking Power on De Novo Binders
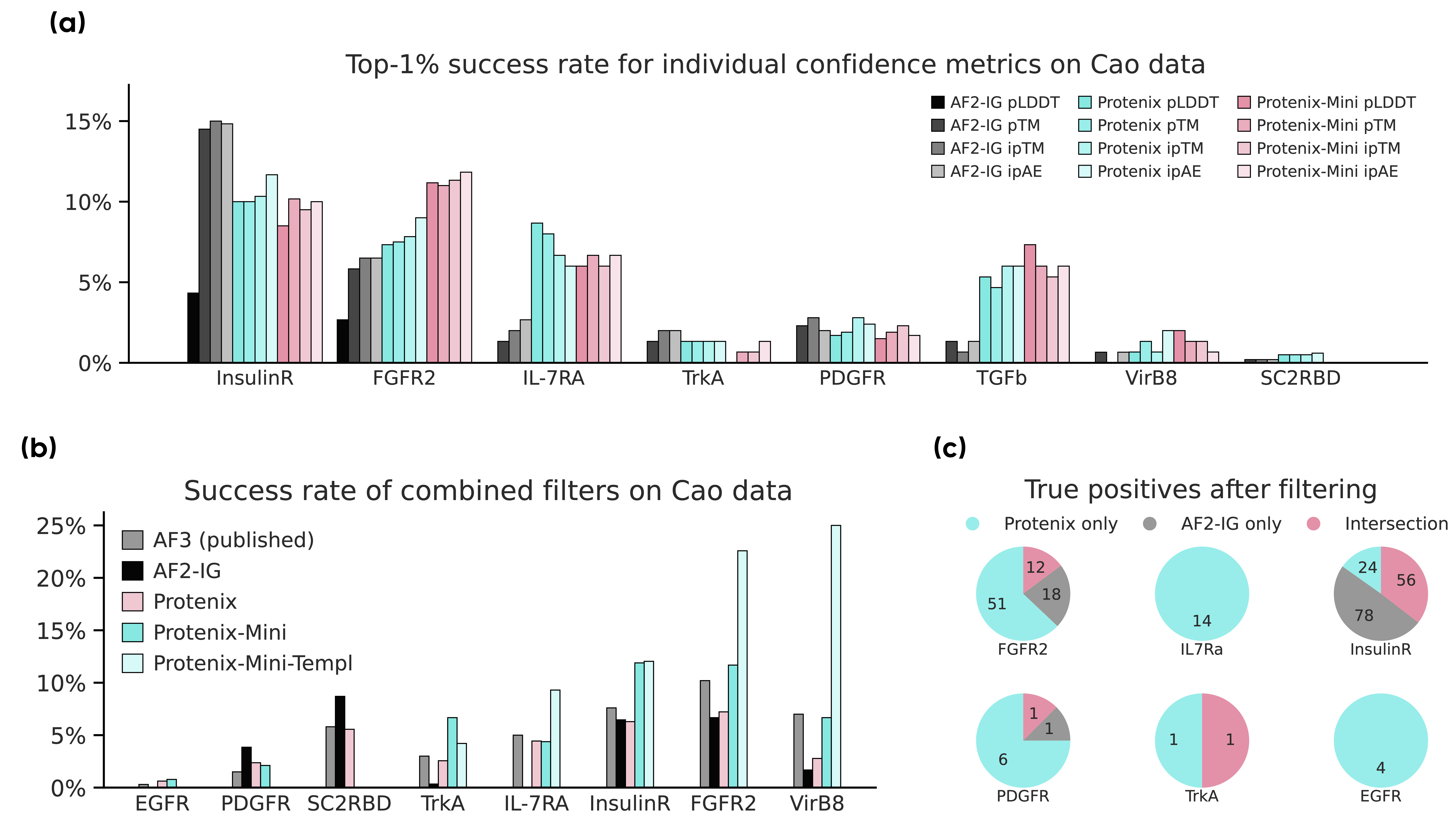
Protenix-based filtering strongly enriches and prioritizes true binders;
together with AF2-IG, it captures complementary true positives,
and using both is likely to yield stronger enrichment.
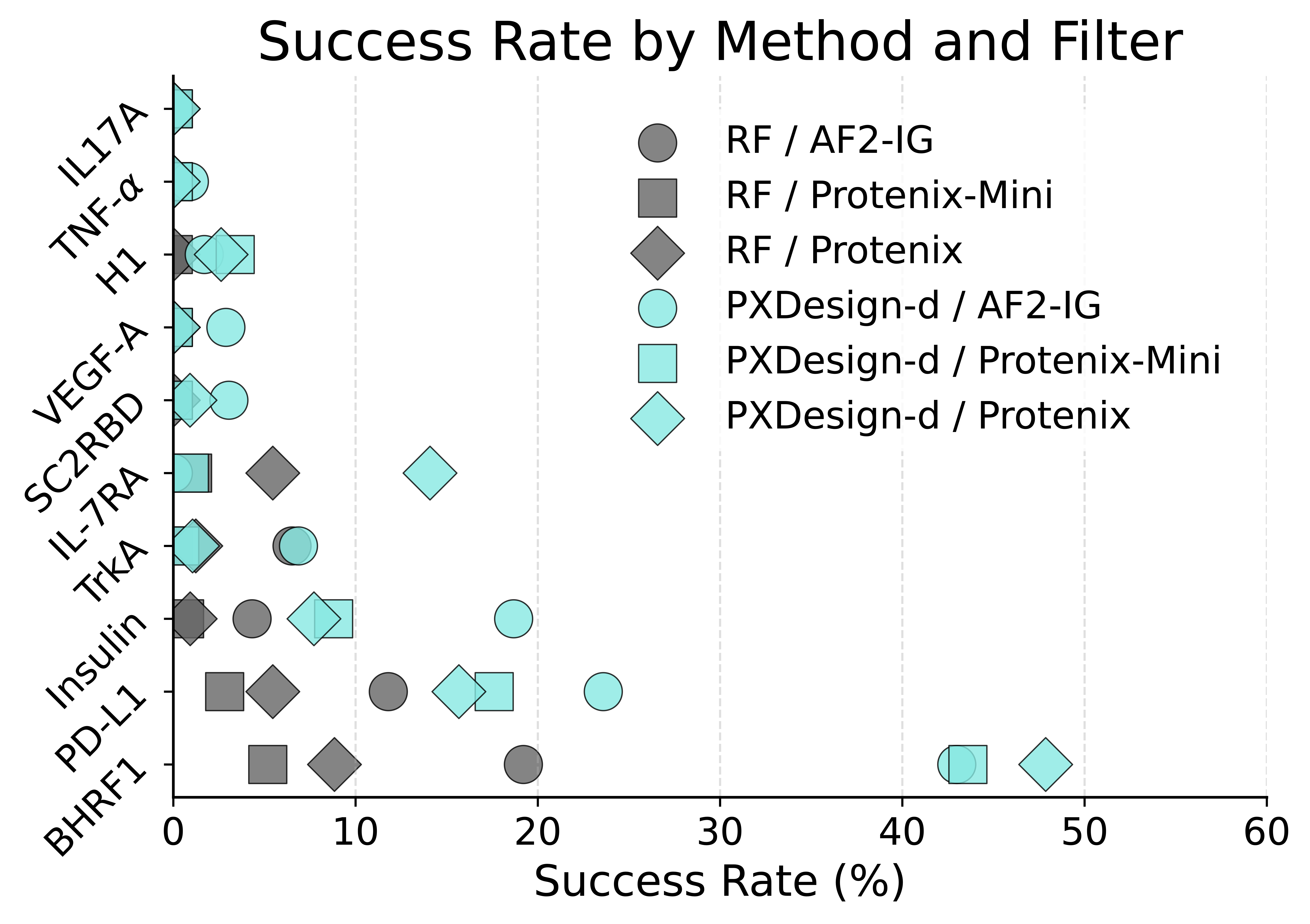
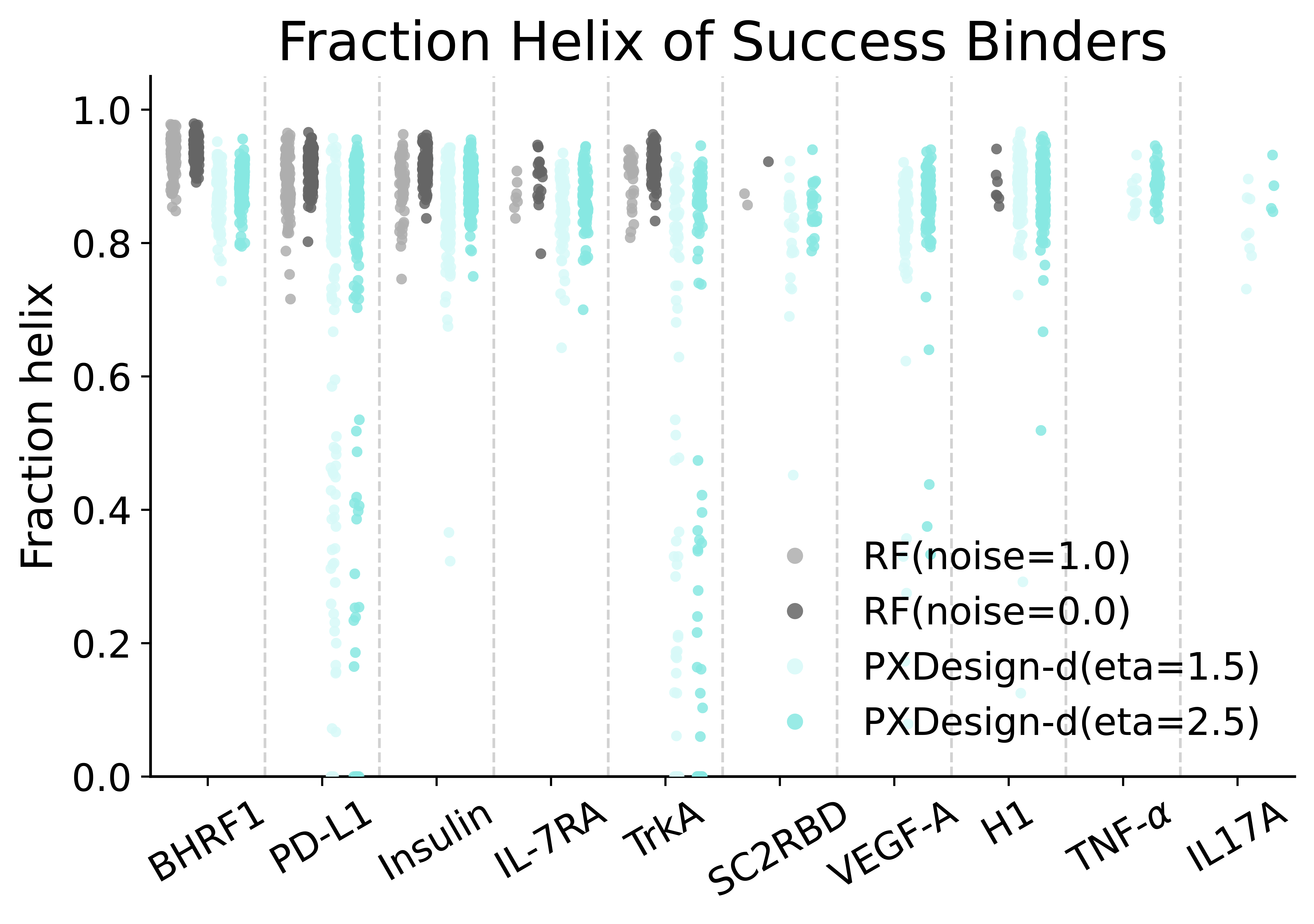
4) In-silico Success Rate of De Novo Binder Generation
PXDesign attains higher success rates and broader fold diversity than RFDiffusion on 10 targets; diffusion is also more throughput-efficient than hallucination for large campaigns.
PXDesign Server Walkthrough
Beyond Protein Binders
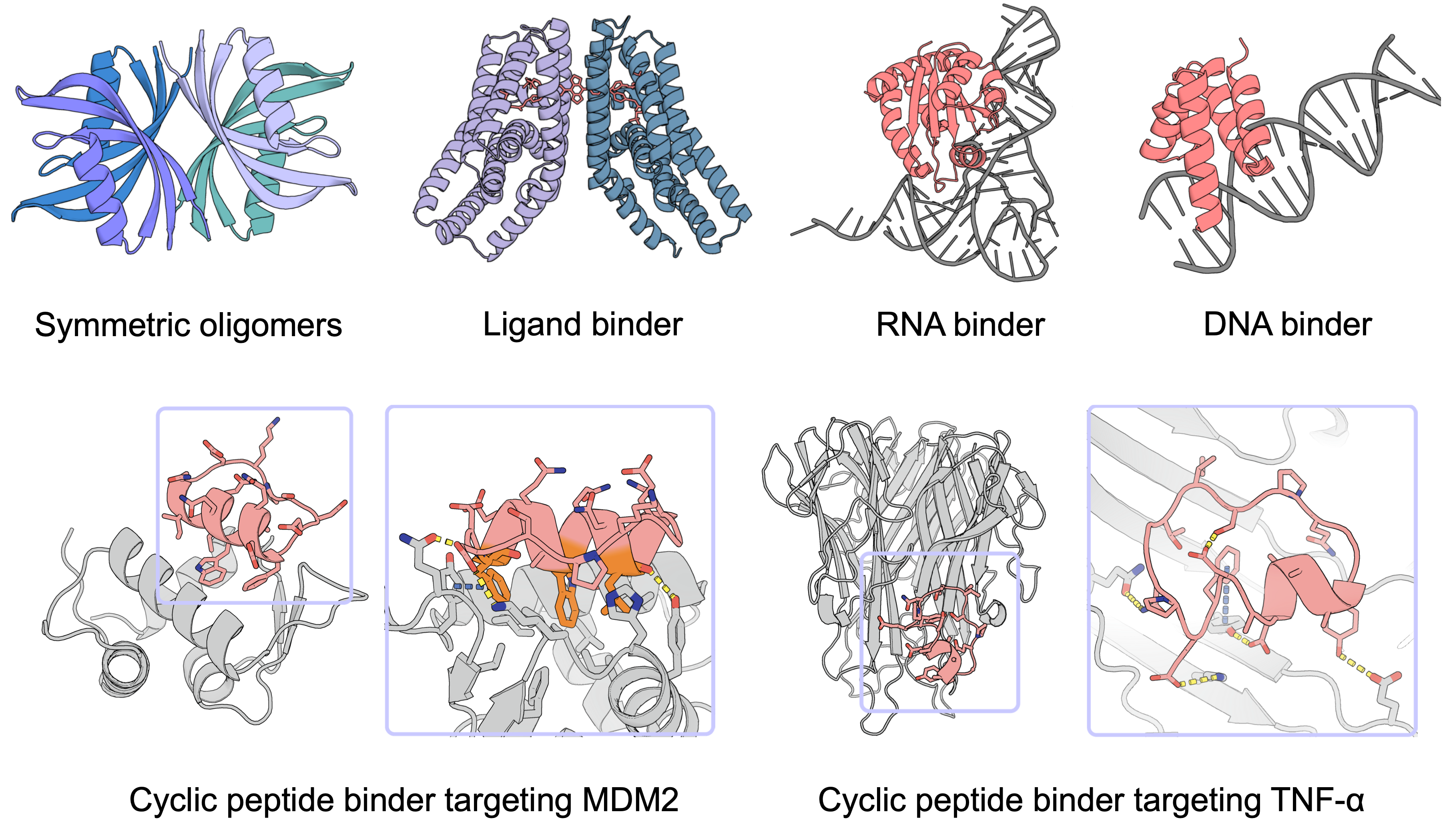
While our in-silico and wet-lab validation focuses on protein binders, PXDesign is naturally extensible to diverse molecular targets (e.g., nucleic acids, small molecules, post-translationally modified proteins).
Ship Notes & Key Takeaways
What’s released
Hosted access:
Publicly accessible Protenix Web Server (for structure prediction) and PXDesign Web Server (for binder design) are available for immediate use.
Foundation Models & Tooling:
Protenix Github repository: full open-source release of the Protenix foundation model.
PXDesign Github repository: full open-source release of the PXDesign model and design pipeline.
Research Assets:
Technical reports for both Protenix and PXDesign, including protocols, thresholds, datasets, and methodology details to support reproducible research.
Key Takeaways
Strong performance from structure prediction to design:
Protenix delivers accurate structural inference, while PXDesign builds on this foundation with diffusion-based generation and orthogonal filtering — yielding high hit rates and diverse binders across multiple targets.
Ready for real workflows:
Both models and tooling are released open-source and ready-to-use. Open benchmarks and public servers make it easy for researchers to reproduce, validate, and build upon the entire Prediction & Design pipeline.